What’s the best way to do these kinds of groupings?

What not to do
Let’s start by putting one idea to rest. You might be tempted to address the roll-up challenges upfront, by mapping your source data to the most generic LOINC code possible.
Mapping to only unspecified specimen and methodless terms seems at first like a good way to simplify your mapping and get broader data buckets for reuse. Here’s my warning:
Don’t ever “throw away” information when mapping
As I mention in my Top 10 Tips for Mapping to LOINC, you’ve got to watch out for the unidirectional effect of data consolidation. Once you assign a more general mapping, you can’t easily go back. I always recommend that you map to the most specific LOINC code that matches the level of fidelity you have in the local concept.
If you preserve the level of granularity present in the local term when mapping to LOINC codes, you can use several different techniques to do roll-ups in various ways. This is a much more robust approach.
Before we dive in though, I want to offer two other caveats.
First, as Stan Huff likes to say, if you’ve seen one hierarchy, you’ve seen one hierarchy. There are often very similar, but different perspectives on how to construct these things and what you might find useful. The techniques you might choose will depend on your use and purpose.
Second, you have to know what you’re doing. Obvious, but I need to say it. Just because you can group similar LOINC codes doesn’t mean that you should.
All approaches should be validated before use in any aspect of clinical care.
HbA1c as a warning
Let me give a quick example of why you need to pay attention whenever you are looking to group results from different LOINC terms. People have often queried LOINC and noticed several codes for Hemoglobin A1C. Naturally, they ask, how can I get “all” the HbA1c terms?
Well, the reason there are multiple terms in LOINC is that there are multiple standardization protocols for measuring Hgb A1c:
- IFCC: the Reference Method
- NGSP: long standing protocol used in the US and most other countries
- JDS/JSCC: a protocol used in Japan, Spain and possibly other countries
- Swedish: used in Sweden at least
The issue is that protocols produce different results, even when expressed in the same units. There’s a nice summary of the relationship of HbA1c results by different protocols on the NGSP site. But, for example, the equivalent of Hgb A1c (NGSP) of 6.5% is Hgb A1c (IFCC) of 4.8%.
In other words, there is a 30% difference between the values produced by these two HbA1c protocols!
You definitely don’t want to blindly combine result values produced by different HbA1c protocols. And if you did want to make comparisons, you’d have to use validated conversion techniques.
Many valid uses for grouping data coded with LOINC
Ok, you’ve heard my main caution: be mindful of your purpose for aggregation and the clinically-important differences between test results identified by different LOINC codes.
Building a research cohort with all the patients who have had a “sodium” measure is different from aggregating or plotting those measures together (ignoring the differences between sodium measures made on stool vs urine vs blood vs serum, etc). For example, sodium levels in preterm infants simultaneously determined using an arterial blood gas analyzer (direct method on undiluted whole blood) and an autoanalyzer (indirect method on diluted plasma) are significantly different.[1]Still, there are lots of valid reasons why you may want to retrieve or aggregate data identified with different LOINC codes that could be considered “equivalent” for your purpose.
Here I’ll present four techniques for rolling up LOINC codes into groups.
(Psst…the newest, hottest one is up first).
1. Use the brand new LOINC Groups
With funding support from the National Center for Advancing Translational Sciences, the LOINC team at Regenstrief has launched an exciting new project to address this specific issue. We’re calling the exciting new project: LOINC Groups.
The LOINC Groups project aims to provide a flexible, extensible, and computable mechanism for rolling up groups of LOINC codes for various purposes. LOINC Groups are designed to capture “equivalence classes”, or areas where certain sets of LOINC codes could be reasonably aggregated together for clinical flowsheets, and perhaps other purposes like decision support.
The first set of LOINC Groups was published with the LOINC 2.61 Release in June 2017, but this project is a work in progress. The initial release contained 2,178 Groups (organized into 12 top level categories called Parent Groups) that organized a total of 6438 unique LOINC terms.
I’d encourage you to download the file and check them out.
The Regenstrief’s team’s approach to creating these LOINC Groups is pretty novel. Of course, we are informed by the existing ontologic relationships that already exist in the current Multiaxial Hierarchy and available through SNOMED CT (both discussed below). But, this effort is focused on clinically-relevant aggregations and not solely ontologic relationships between attributes (LOINC Parts) used in the LOINC terms. A hierarchy like this does not currently exist in a mature, scalable, and sustainable form.
For example, existing relationships can determine that venous blood specimen and arterial blood specimen are both subsumed by blood specimen. But tests on other kinds of blood specimens, like cord blood and dried blood spots, have very different uses clinically. Tests on these specimens would not be plotted together on the same line of a flow sheet. If you only traversed ontological relationships, you would follow the path that included cord blood, because sodium measured in cord blood is_a sodium measured in blood. While true, this isn’t very useful clinically, so in the LOINC Group we kept them out:
Example LOINC Group for Sodium
[LG11309-8] Sodium|SCnc|Pt|ANYBldSerPlas
[2947-0] Sodium [Moles/volume] in Blood
[39792-7] Sodium [Moles/volume] in Capillary blood
[41657-8] Sodium [Moles/volume] in Mixed venous blood
[2951-2] Sodium [Moles/volume] in Serum or Plasma
[77139-4] Sodium [Moles/volume] in Serum, Plasma or Blood
[39791-9] Sodium [Moles/volume] in Venous blood
As you can see from the syntax of the Group name, this collects all LOINC terms that have Component of Sodium, Property of SCnc, and Timing of Pt; ANY blood, serum or plasma System (with the exceptions like cord blood and dried blood spot); and ANY Method.
An ongoing work in progress
Regenstrief’s approach to creating LOINC Groups is being designed to take into account test characteristics and clinical interpretation issues that influence how or why users would make use of them. The plan is to not only to extend the scope of LOINC Groups created, but to also extend the metadata to support annotated usage notes along with the computable structures. We took a similar approach with the subset of the most common laboratory result codes (dubbed the Top 2000) that also included mapping guidance for users.
Annotations in the LOINC Groups aggregation hierarchy will help inform users when a particular grouping may be applicable. For example, an aggregation of all LOINC codes for a given analyte performed on various blood subtypes (arterial, venous, capillary, cord blood, etc) would be helpful in many circumstances. But, what if there is a documented 7% difference in the arterial versus venous measures? Should you aggregate those results together? Of course, it depends on your purpose. Annotations will help users make an informed choice.
Regenstrief also plans to extend and refine the approach to documenting molecular weights and other conversions.
Feedback welcome
The Regenstrief team is very interested in hearing your feedback on the current file format and the LOINC Groups that have been created. I strongly recommend downloading the file, exploring, and then posting comments over on the LOINC forum.
2. Use the LOINC Multiaxial Hierarchy File
Regenstrief also produces a “multi-axial” hierarchy file that provides a way to organize LOINC codes based on multiple axes of each concept. This file is distributed with each LOINC release, and organizes lab tests first by Component, then by System. LOINC Parts comprise all of the branches in the Multiaxial Hierarchy, and it groups LOINC codes under those branches as leaf nodes.
The Multi-axial Hierarchy is generated using an automated process based on the individual Class, Component, System, and Method hierarchies, which are maintained by manually by the LOINC staff at Regenstrief. These hierarchies are not meant to be pure ontologies, and rather are tools for organizing sets of codes within different domains. It can be quite useful for finding siblings, etc.
The LOINC Multiaxial Hierarchy File is available for download as a CSV file, and again, I’d highly encourage you to read the MULTI-AXIAL_HIERARCHY_Readme.txt file to get your bearings.
You can also browse and search the LOINC Multiaxial Hierarchy in RELMA. Head over to the “Hierarchy & Search Limits” tab, then choose “Multi-axial Hierarchy”. There is a search bar at the top where you can enter key words:
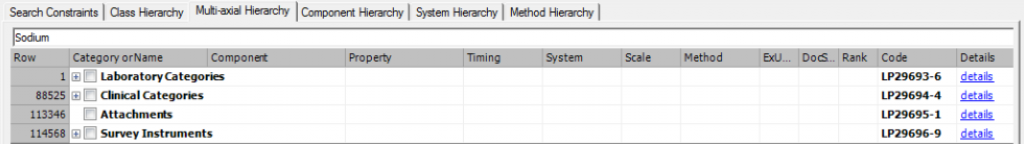
After executing a search, click the “Show LOINCs” button to display the LOINC codes under a particular branch. Here are the terms nested under the Part LP48861-6 Sodium | Bld-Ser-Plas:
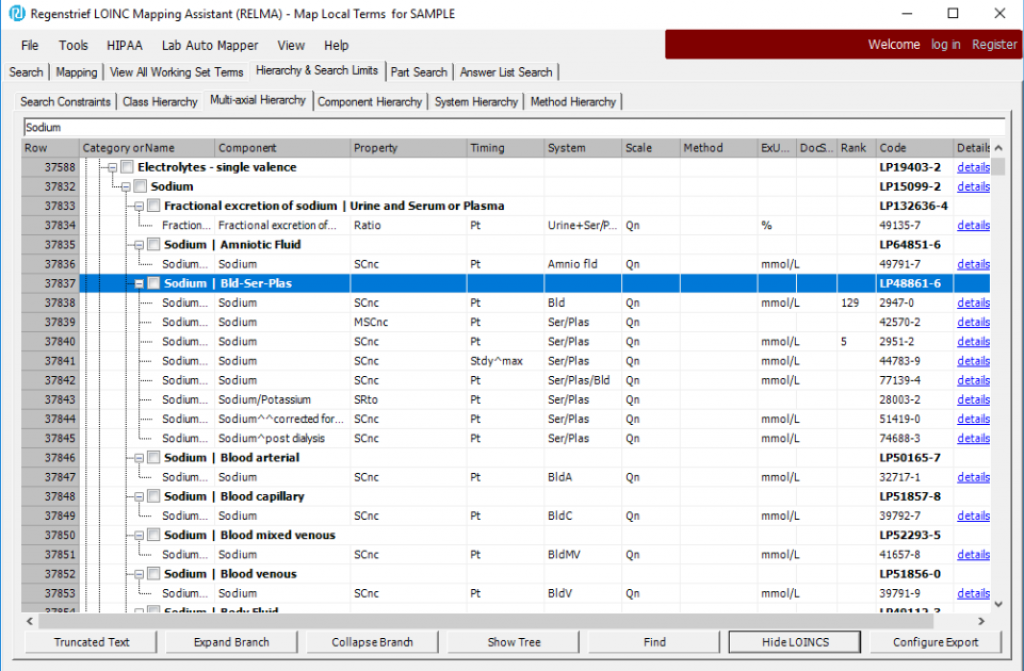
RELMA is a great way to visualize the hierarchy, and you can can select rows to export from this screen too.
The LOINC Multiaxial Hierarchy File CSV format is better for programmatic queries and loading into a terminology server. To locate all of the LOINC codes nested under the branch of Part LP48861-6 Sodium | Bld-Ser-Plas, just look for the rows where the IMMEDIATE_PARENT is LP48861-6:
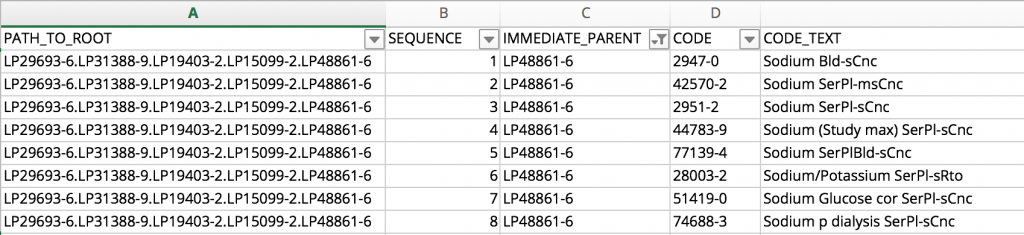
Whether or not this collection of Sodium terms is the exact collection of LOINC codes you need will depend on your use case, but the Multiaxial Hierarchy is useful for illustrating an overall organization of LOINC codes and grouping them according to key parts of the name.
3. Use the established linkages between LOINC and SNOMED CT
Since 2013, Regenstrief has been working closely with SNOMED CT (formerly known as IHTSDO) under our long term collaborative agreement. The first sets of artifacts from that collaboration include two main kinds of linkages between the terminologies.
One is a mapping of LOINC part codes to SNOMED CT concepts. These mappings are made available both in SNOMED’s LOINC Part to SNOMED CT Map Reference Set format (SNOMED RF2) and as a component of the LOINC Part File, which includes a PartRelatedCodeMapping_Alpha_1.csv table that has the linkages.
The other artifact produced from this collaboration is a linkage between LOINC observation codes and a SNOMED CT expression that models that LOINC concept according to the new (still in progress) SNOMED CT Observables model. At this point, the collaborative work covers a subset of LOINC codes, but is expanding as we get resources and time.
From these files you’ll be able to use whichever SNOMED CT relationships/hierarchies you find useful for organizing the LOINC codes. Before diving in, I’d definitely recommend reviewing the Release Notes accompanying the material from SNOMED International and the relevant Section in the SNOMED CT Editorial Guide (6.1.3.2 Observable Entities and Evaluation Procedures) that explains the attributes of the SNOMED CT observables model.
Let’s suppose that you would like a query to answer the question whether your patient has had an HIV Ab or Ag test. You might start by looking for SNOMED CT expression associations containing:
Lab tests with Component:
Human immunodeficiency virus antibody [259855002] OR Human immunodeficiency virus antigen [116982009] OR Child concepts of either
To find the terms you want, the hierarchical relationships in SNOMED CT followed down the tree and used used to organize the terms by HIV subtype.
A snippet of the SNOMED CT hierarchy showing LOINC terms with HIV 1 Components :
Human immunodeficiency virus antibody [259855002]
Human immunodeficiency virus type 1 antibody [120841000]
Human immunodeficiency virus 1 protein 24 antibody [444013004]
[16978-9] HIV 1 p24 Ab [Units/volume] in Serum
[43011-6] HIV 1 p24 Ab [Presence] in Serum
[35448-0] HIV 1 p24 Ab [Presence] in Saliva (oral fluid) by Immunoblot
…
Human immunodeficiency virus 1 protein 68 antibody [445463006]
[12894-2] HIV 1 p68 Ab [Presence] in Serum by Immunoblot
…
4. Use direct queries on the LOINC database
LOINC names are created using structured attributes and controlled attribute values called LOINC Parts. So, within RELMA or the LOINC Table itself, these attributes can help you find all measures of a particular analyte on a given specimen type.
Using RELMA
In RELMA, you can use the Google-like search syntax to query for keywords within specific LOINC attributes. This search finds LOINC terms with Sodium in the Component and Ser in the System:
component:sodium system:ser
RELMA uses its keywords and synonyms to return these LOINC terms:
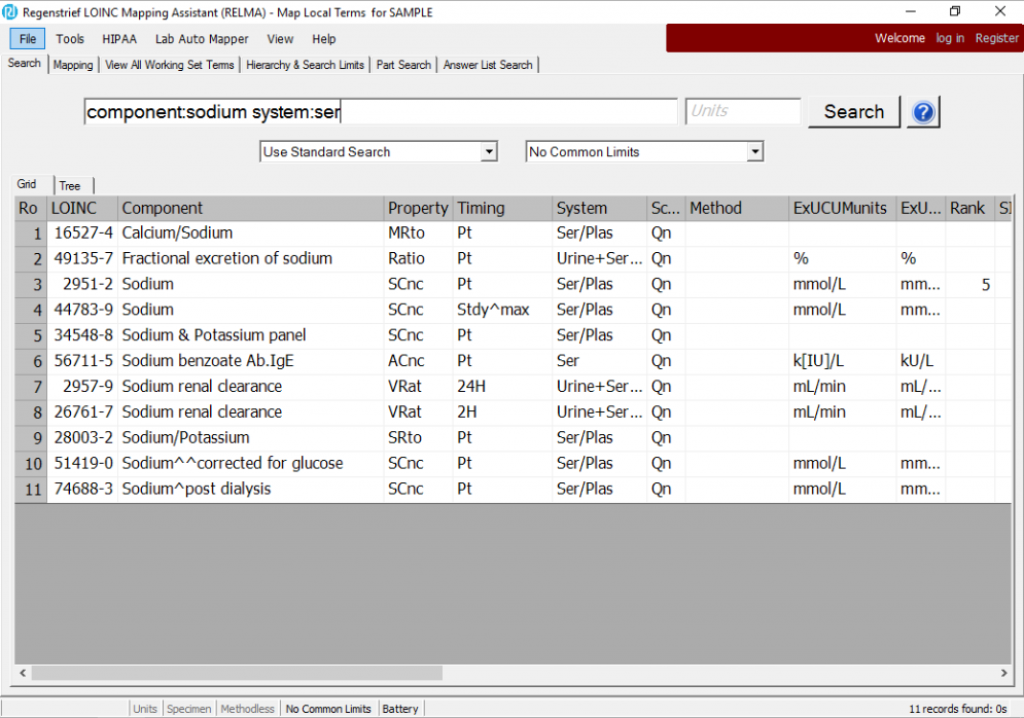
Looking at this list, these terms are perhaps more of a mixed bag than you would have expected. There are ratios, panels, and other variants you might not have been expecting. To narrow that down a little bit, you’ll need more specificity in your search:
component:sodium system:ser scnc -panel
This search returns four LOINC terms that you might consider “equivalent” for certain purposes.
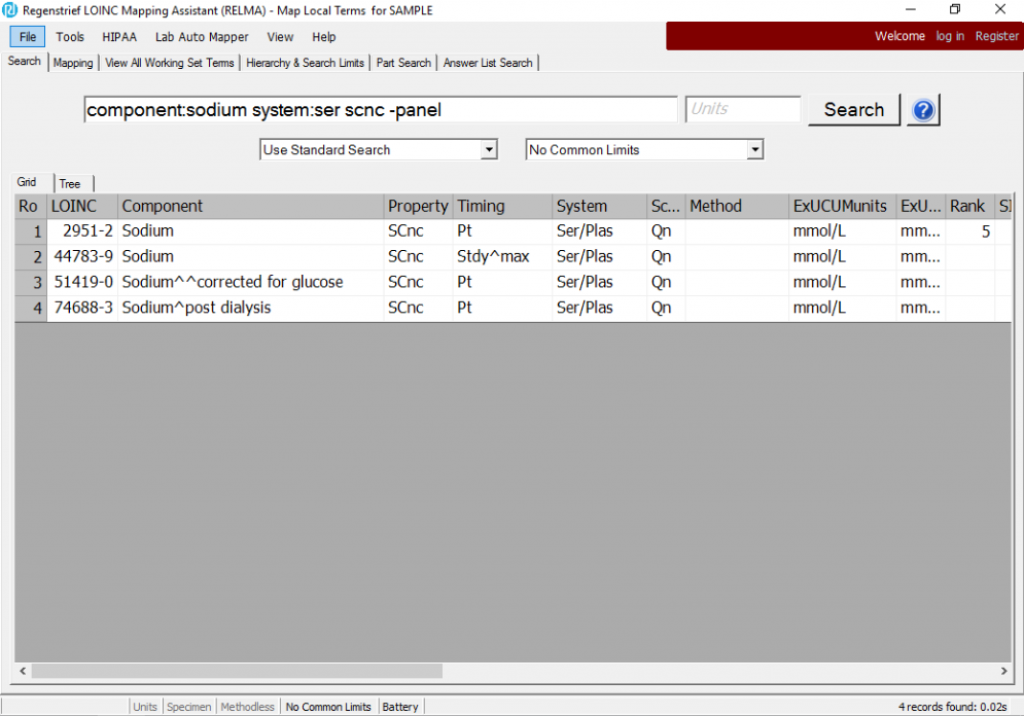
Once you have narrowed your search down to the terms you want in your group, simply select the rows and Export to the clipboard, a CSV file, or Excel. (RELMA’s configurable export feature lets you choose what elements from the LOINC record you want in the export, e.g. LOINC code, Long Common Name, etc).
Using SQL on the LOINC table itself
The LOINC Table has fields for each of the main LOINC name attributes (as well as many other metadata attributes). With SQL, you can select the relevant LOINC codes based by querying these attributes.
Here is a query for returning all LOINC terms with Status of Active, Component of Sodium, and System of Ser/Plas:
SELECT LOINC.LOINC_NUM, LOINC.LONG_COMMON_NAME
FROM LOINC
WHERE (((LOINC.COMPONENT)="SODIUM") AND ((LOINC.SYSTEM)="Ser/Plas") AND ((LOINC.STATUS)="ACTIVE"));This query returns the following terms:
[2951-2] Sodium [Moles/volume] in Serum or Plasma [44783-9] Sodium [Moles/volume] (Maximum value during study) in Serum or Plasma
What happened to 51419-0 and 74688-3?
Notice that the query above uses an exact match on Component of Sodium and not a “contains” or “begins with”. Recall that the Challenge LOINC Subpart is also part of the LOINC Table field COMPONENT (separated by a “^” character). If we want to include Sodium challenge tests, we’d need to add an option to find them.
To be sure we were only getting substance concentration terms and no panel codes, we could add criteria for that too:
SELECT LOINC.LOINC_NUM, LOINC.LONG_COMMON_NAME, LOINC.PROPERTY, LOINC.PanelType
FROM LOINC
WHERE (((LOINC.COMPONENT) Like "SODIUM*") AND ((LOINC.PROPERTY)="SCnc") AND ((LOINC.SYSTEM)="Ser/Plas") AND ((LOINC.STATUS)="ACTIVE") AND ((LOINC.PanelType) Is Null));
Now, we’re back to the same four terms we saw earlier:
[2951-2] Sodium [Moles/volume] in Serum or Plasma [44783-9] Sodium [Moles/volume] (Maximum value during study) in Serum or Plasma [51419-0] Sodium [Moles/volume] corrected for glucose in Serum or Plasma [74688-3] Sodium [Moles/volume] in Serum or Plasma --post dialysis
Using the new LOINC Part File
New with LOINC version 2.61 was an additional artifact that links LOINC terms to their associated Parts. This artifact makes it possible to use the actual LOINC Part Codes (LP*) in your queries. Be sure to take a look at the PartFile_Readme.txt file to familiarize yourself with the structure and contents of these tables. You’ll want to join these tables with the main LOINC Table to have all the attributes at your disposal for queries.
Here’s just a simple example that returns all of the LOINC terms (including deprecated ones) linked to LP15099-2 Sodium with a LinkTypeName of Primary:
SELECT DISTINCT LoincPartLink_Alpha_1.[LoincNumber], LoincPartLink_Alpha_1.LongCommonName
FROM LoincPartLink_Alpha_1 INNER JOIN Part_Alpha_1 ON LoincPartLink_Alpha_1.PartNumber = Part_Alpha_1.[PartNumber]
WHERE (((Part_Alpha_1.[PartNumber])="LP15099-2") AND ((LoincPartLink_Alpha_1.LinkTypeName)="PRIMARY"));
This query includes some molar ratio and other terms you’d likely not want to include in your roll-up. To filter them out you’d need a more advanced query.
Wrap up
We’ve covered four different techniques for rolling up LOINC codes into higher level groups. Which one you use may depend on your specific use case. I highly encourage you to take a look at the new LOINC Groups and give your feedback. In my opinion, these are going to be a huge boon to users looking to equivalence and aggregate data coded with LOINC.
Acknowledgments
This material contains content from LOINC® (http://loinc.org). The LOINC table, LOINC codes, and LOINC panels and forms file are copyright © 1995-2017, Regenstrief Institute, Inc. and the Logical Observation Identifiers Names and Codes (LOINC) Committee and available at no cost under the license at http://loinc.org/terms-of-use.

